Matrix-Assisted Laser Desorption/Ionization
Matrix-Assisted Laser Desorption/Ionization (MALDI) is a soft ionization technique used in Mass Spectrometry to analyze larger molecules, which are either non-volatile or thermally unstable. This technique allows for identification and spatial distribution studies of biomolecules (e.g. DNA, proteins, peptides and sugars) and large synthetic organic molecules (e.g. polymers, dendrimers and other macromolecules).
MALDI is a unique technique allowing for desorption and ionization of entire molecules, unlike more conventional ionization techniques, which often lead to substantial fragmentation of the species of interest. Initially, the sample is mixed with a selected matrix, such α-cyano-4-hydroxycinnamic acid, and deposited on a target plate. This plate is bombarded by photons from a pulsed laser, resulting in the desorption and ionization of the matrix. Subsequently, the energy is transferred from the matrix to the sample molecules. This gentle energy transfer process leaves the sample molecules intact, but now in the gas phase, yielding protonated/ cationized or deprotonated/anionized molecular ions.
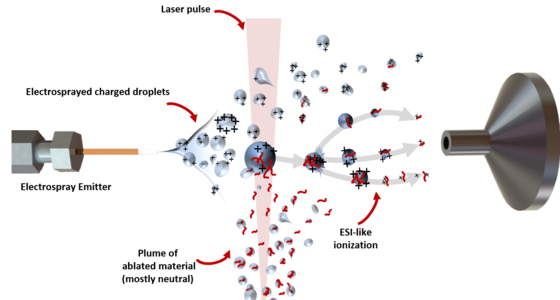
The ions are analyzed with a Time-of-Flight Mass Spectrometer (ToF MS). Using this technique, the mass of the ions is determined by separation of the ions in time according to their mass/charge (m/z) ratio. Scans of the whole mass range are taken in ToF MS to obtain a full mass spectrum. ToF MS enables one to determine the molecular masses of the ions with high accuracy. In most cases, obtaining the mass of a specific molecule is not sufficient for unique identification. A route to acquire this information is tandem mass spectrometry (MS/MS or ToF/ToF). First, a selected ion is isolated and subsequently fragmented. The parent ion and fragment ions are then separated in the second ToF mass spectrometer, yielding a pattern of fragments, forming a characteristic fingerprint of the molecule of interest.
Ideal Uses of MALDI
Accurate molecular weight measurements
- Sample identification
- Determination of the purity of a sample
- Verification of amino acid substitutions
- Verification of post-translational modifications
Reaction monitoring
- Enzyme reactions
- Chemical modification
- Protein digestion
Amino acid and oligonucleotide sequencing
- Confirmation of amino acid sequence
- De novo characterization of peptides
- Identification of proteins by database searching with a sequence “tag” from a proteolytic fragment
- Characterization or quality control of oligonucleotides
Protein structure
- Protein folding monitored by H/D exchange
- Protein-ligand complex formation under physiological conditions
- Macromolecular structure determination
Bacterial and fungal identification
- Identification of bacterial and fungal isolates
- Spatial distribution of compounds within biological tissues or non biological systems
Strengths
- Rapid, sensitive, high mass accuracy, high throughput
Limitations
- Soft ionization not suitable for all molecules. Mass discrimination for wide polydispersity polymers
MALDI Technical Specifications
Information obtained:
- Composition, molecular weight and structural information on individual components
- Qualitative and (semi-)quantitative
- Sample type: Solid or liquid (dissolved in aqueous buffers or solvents such as DMF or DCM)
Mass range: 50 – 100000 g/mol
Mass accuracy:
- 0.1% (1000 ppm) for masses
- > 4000 g/mol
- 0.005% (50 ppm) or better for masses
- < 4000 g/mol
- 50×10-3 g/mol in MS/MS mode
Mass sensitivity: Compound and matrix dependent
Lateral resolution (imaging): 20 x 20 μm
The ions are analyzed with a Time-of-Flight Mass Spectrometer (ToF MS). Using this technique, the mass of the ions is determined by separation of the ions in time according to their mass/charge (m/z) ratio. Scans of the whole mass range are taken in ToF MS to obtain a full mass spectrum. ToF MS enables one to determine the molecular masses of the ions with high accuracy. In most cases, obtaining the mass of a specific molecule is not sufficient for unique identification. A route to acquire this information is tandem mass spectrometry (MS/MS or ToF/ToF). First, a selected ion is isolated and subsequently fragmented. The parent ion and fragment ions are then separated in the second ToF mass spectrometer, yielding a pattern of fragments, forming a characteristic fingerprint of the molecule of interest.
Ideal Uses of MALDI
Accurate molecular weight measurements
- Sample identification
- Determination of the purity of a sample
- Verification of amino acid substitutions
- Verification of post-translational modifications
Reaction monitoring
- Enzyme reactions
- Chemical modification
- Protein digestion
Amino acid and oligonucleotide sequencing
- Confirmation of amino acid sequence
- De novo characterization of peptides
- Identification of proteins by database searching with a sequence “tag” from a proteolytic fragment
- Characterization or quality control of oligonucleotides
Protein structure
- Protein folding monitored by H/D exchange
- Protein-ligand complex formation under physiological conditions
- Macromolecular structure determination
Bacterial and fungal identification
- Identification of bacterial and fungal isolates
- Spatial distribution of compounds within biological tissues or non biological systems
Strengths
- Rapid, sensitive, high mass accuracy, high throughput
Limitations
- Soft ionization not suitable for all molecules. Mass discrimination for wide polydispersity polymers
MALDI Technical Specifications
Information obtained:
- Composition, molecular weight and structural information on individual components
- Qualitative and (semi-)quantitative
- Sample type: Solid or liquid (dissolved in aqueous buffers or solvents such as DMF or DCM)
Mass range: 50 – 100000 g/mol
Mass accuracy:
- 0.1% (1000 ppm) for masses
- > 4000 g/mol
- 0.005% (50 ppm) or better for masses
- < 4000 g/mol
- 50×10-3 g/mol in MS/MS mode
Mass sensitivity: Compound and matrix dependent
Lateral resolution (imaging): 20 x 20 μm
